Use DSC to follow the stability of antibodies during the critical steps of manufacturing. There are multiple challenges to overcome while developing formulations for proteins intended as therapeutics. Most notably, project timelines and often limited initial availability of material drive the need for rapid, robust, and sensitive formulation development tools. Thermodynamic analysis has proven to be an invaluable approach to aid in the characterization of physical protein instabilities. However, the generated thermodynamic parameters, such as melting temperature (Tm), are typically apparent rather than absolute. Even with this caveat, apparent thermodynamic parameters are indispensable in rapidly evaluating protein instabilities. Differential Scanning Calorimetry (DSC) is a key tool in the analytical arsenal of a formulation development scientist. DSC allows for the determination of calorimetric melting temperatures and enthalpies of unfolding that permit an assessment of protein solution behavior under a variety of conditions. Several examples of pre-formulation and later-stage formulation applications of DSC, as well as their limitations and other special considerations, are discussed. These examples illustrate the crucial role of DSC and thermodynamic analysis during therapeutic protein development.
There are many challenges to overcome when developing manufacturing processes for proteins intended as therapeutics. Most notably, project timelines and limited initial availability of material drive the need for rapid, robust, and sensitive process development tools. Manufacturing steps are routinely evaluated and optimized to improve protein yield, reduce production time, and increase long-term stability of final drug product. Any improvements in final yield and stability are vastly important in both process and manufacturing economics. Conversely, a failure at any step in production, resulting in decreased yield and/or stability, will result in a financial loss.
Thermodynamic analysis has proven to be an invaluable approach to aid in the characterization of physical protein instabilities. Understanding the factors which affect stability, such as pH, ionic strength, and excipients, are critical in the design and optimization of manufacturing processes.
Differential Scanning Calorimetry (DSC) is a powerful analytical tool to assess these stability parameters. DSC is utilized throughout the biotherapeutic development pipeline, since it quickly and easily facilitates the selection of both process conditions as well as storage conditions during manufacturing. DSC generates thermodynamic parameters, such as melting temperature (Tm), which are indispensable in rapidly evaluating protein stability.
In this application note, based on a recent presentation by Genzyme scientists, we report the use of the thermal stability data obtained from DSC to screen conditions for viral inactivation of an antibody1. This information provides a quick and easy stability assay during the manufacturing process. For the antibody studied here, the apparent Tm data from DSC was key to the recommendation to increase storage pH immediately after viral inactivation, resulting in increased structural stability. This example illustrates the crucial role of DSC and thermodynamic analysis during therapeutic protein development.
The ultimate goal of the development of proteins as therapeutics is the design of dosage forms with sufficient stability to withstand shipping, processing, and storage stresses. Biopharmaceutical development is complex and requires expertise and input from multiple functional areas, such as process development, manufacturing, and formulation development. Usually the allotted time is compressed and real-time stability assessments for the shelf-life of the dosage forms are impractical. Therefore, bioanalytical scientists need to rely on a combination of accelerated and forced degradation studies to probe the likely instabilities that might be encountered for a given protein. Methods that help provide an early assessment of the solution behavior of a protein are very valuable. Nevertheless, the amount of material available for analysis during early stage evaluations is often a limiting factor. Procedures that employ small quantities of protein and provide information regarding structure and stability are essential. An initial thermodynamic evaluation of a protein resulting from these procedures can provide insight into its solution behavior under a variety of conditions. A preferred tool for such analysis is Differential Scanning Calorimetry (DSC). Although information could also be obtained by alternative methods, DSC has several key advantages: high throughput capabilities (using the automated VP- Capillary DSC system), reproducibility, virtually unlimited buffer compatibility, and most importantly, it is a first-principle technique where the enthalpy of a given reaction can be measured directly.2-4
Instabilities most commonly observed during development of biotheraputic proteins fall into two categories: biochemical and structural. Some of the most common chemical instabilities include oxidation, deamidation, and isomerization.5-9 Characterization of chemical instabilities typically relies on forced degradation studies that utilize temperature or biochemical stresses to identify relevant degradation pathways.7, 10, 11 Altering protein chemical stability is accomplished by changing solution conditions based on knowledge of the chemistry underlying the instability of interest. Additional degradation pathways observed include: secondary and tertiary structure changes, fragmentation, and aggregation/self-association. 7, 12, 13
The application of thermodynamics to manufacturing development is typically governed by three key stabilization mechanisms: 1) minimization of partially unfolded intermediates prone to aggregation, 2) stabilization of the folded state, leading to aggregation prevention, through the preferential exclusion mechanism, and 3) structural stabilization by maximizing protein-ligand or proteinexcipient binding interactions. Maximizing thermodynamic stability can also indirectly impact biochemical stability by changing the solvent exposure of susceptible residues. There are several challenges that may be encountered when applying thermodynamics to process development and manufacturing issues. First, project timelines often demand the ability to perform high-throughput screening characterization. Second, the thermodynamic parameters obtained need not be, nor will they be, absolute; they are apparent parameters, useful for relative comparisons of manufacturing conditions. The inability to calculate absolute thermodynamic parameters is a consequence of the irreversible unfolding behavior typically observed in large, multi-domain proteins which often contain disulfide bonds, glycosylation sites, and other post-translational modifications. Finally, thermodynamics provides no direct link to kinetics. During development of a drug with an intended shelf-life, it is critical to understand when and how fast a given reaction will take place. Given those basic precautions, thermodynamic analysis can provide a researcher with valuable information about the relative differences in stability of a given set of solution conditions. In this application note we report the use of the thermal stability data obtained from DSC to screen conditions for viral inactivation of an IgG.
Differential Scanning Calorimetry (DSC): DSC was performed using a high throughput MicroCal VP-Capillary DSC (Malvern Instruments, Northampton, MA). Thermograms for each protein (350 μL at 1 mg/mL) were obtained from 25° C to 100° C using a scan rate of 200° C hr-1, unless otherwise noted. Thermograms of the buffer alone were subtracted from each protein prior to analysis using Origin 7.0 graphing software equipped with the MicroCal VP-Capillary DSC analysis software add-on.
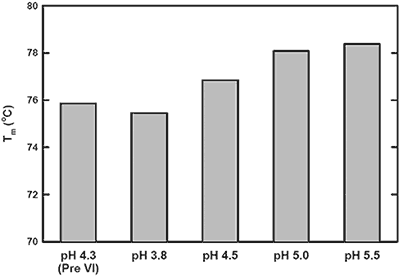
The pH-mediated viral inactivation is a standard industry practice in which the protein is exposed to a decreased pH.14 In this antibody example (antibody-C), increased aggregation rates after viral inactivation were reported by the purification group (data not shown). This observation prompted the use of DSC to characterize the behavior of antibody-C during this processing step, and to subsequently identify an appropriate post viral inactivation storage condition. Samples were exposed to a simulated viral inactivation step, after which the pH was increased in a step-wise manner. The behavior of all three transitions as a function of pH was found to trend consistently. Representative data from a single Tm in Figure 1 shows increased thermal stability with increasing pH. Based on the apparent Tm data, it was recommended to increase storage pH immediately after viral inactivation to maximize structural stability.
|
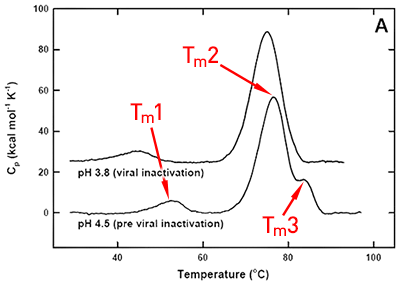
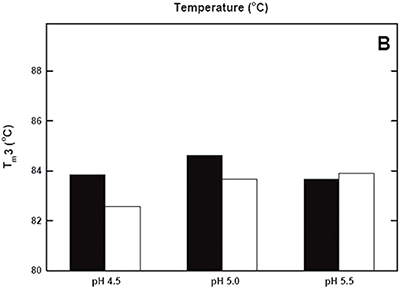
DSC was used not only to improve the process, but also to identify the underlying cause of this instability. Significant destabilization of the first and loss of the third transition is seen when the pH 3.8 thermogram in (Figure 2A) is compared to the pH 4.5 pre-viral inactivation thermogram. Partial loss of antibody-C structure was confirmed by circular dichroism (data not shown), supporting the observation of Fc domain unfolding at low pH. Antibody-C showed decreased thermal stability after viral inactivation (Figure 2B), providing a potential explanation for the increased aggregation rates reported. An analysis such as this can yield a significant amount of valuable information about a protein early in development. It allows for the intelligent design of a viral inactivation strategy and the associated in-process hold steps that minimize protein instability. Additionally, preliminary information about pH and unfolding behavior is collected, which is valuable later informulation development.
|
|
The preceding case study provides one example of the role that thermodynamic characterization and DSC specifically should play in the biopharmaceutical development process. While it is far from an exhaustive list, some other key applications of DSC include: identifying appropriate storage temperatures, characterizing stability as a function of pH and buffer species, enhancing process development activities, and the characterization of stabilizing excipients and ligands during late stage formulation development.
MicroCal thanks Nicholas Guziewicz, Grant Trierweiler, Kendra Johnson, Robert Simler, and Bernardo Perez-Ramirez with BioFormulations Development, Genzyme Corporation, Framingham MA. for contributing their article to the Proceedings of the 2007 Current Trends in Microcalorimetry Conference (reference 1). This application note was adapted from their article.