Prepared by Prof. A. Nakagawa, Institute for Protein Research, Osaka University, Japan, and Kohei Shiba, Sysmex Corporation, Japan
This application note presents the characterization of a virus protein by light scattering to find its size and molecular weight. These parameters may be used to confirm the presence of viral particles and investigate their behavior under different solution conditions.
A typical virus consists of a regular shell of protein molecules, and ranges in size from 20nm (>20 protein units) up to in excess of 100nm diameter (several hundred protein units) [1].
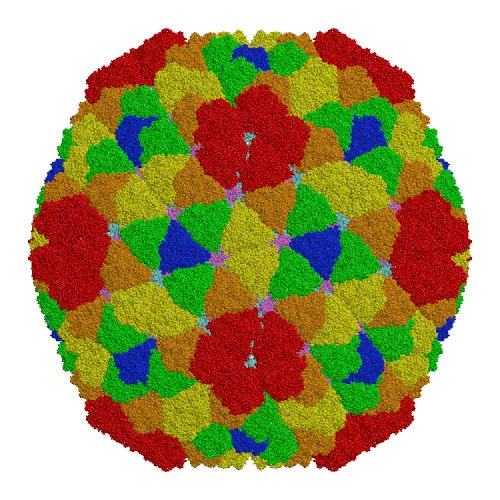
Rice dwarf virus (RDV), the causal agent of rice dwarf disease, is a member of the genus Phytoreovirus in the family Reoviridae and infects rice (Oryza sativa) plants leading to stunted growth, fine chlorotic specks on leaves and subsequent crop damage.[1] RDV is a double-shelled virus with a molecular mass of approximately 70 million Dalton. This virus is widely prevalent and is one of the viruses that cause the most economic damage in many Asian countries. The atomic structure of RDV was determined at 3.5 Å resolution by X-ray crystallography at the Protein Institute of Osaka University [1]. The double-shelled structure consists of two different proteins, the core protein P3 and the outer shell protein P8. The atomic structure shows structural and electrostatic complementarities between both homologous (P3-P3 and P8-P8) and heterologous (P3-P8) interactions, as well as overall conformational changes found in P3-P3 dimer caused by the insertion of amino-terminal loop regions of one of the P3 protein into the other. These interactions suggest how the 900 protein components are built into a higher-ordered virus core structure[1]
|
RDV was cultivated, purified and prepared in water at various concentrations [1]. Two different strains were looked at, RDV+ (which crystallized) and RDV-(which failed to form crystals). All measurements were performed on a Zetasizer Nano ZS at a temperature of 25°C. Samples were measured without any filtration and had low turbidity. The relatively large size of the scattering particles meant that the laser had to be attenuated to avoid saturating the sensitive avalanche photodiode detector. The Zetasizer Nano contains an automatic attenuator over a range of 1:300,000 and is thus able to handle highly scattering samples of large particles without problems.
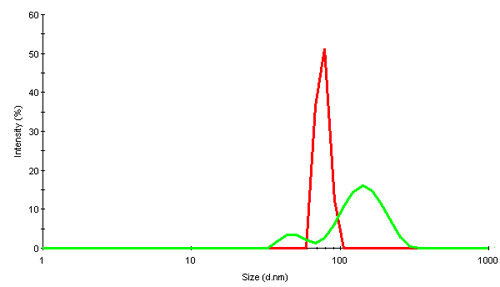
Virus particles are huge assembly complexes of proteins. In dynamic light scattering (DLS), intensity fluctuations in scattered light due to their Brownian motion are analyzed to obtain the diffusion coefficient. Applying knowledge of the viscosity of the diffusing medium (water in this study) a hydrodynamic size of the scattering object can be obtained. The size is that of an equivalent sphere which diffuses with the same diffusion coefficient as the virus. Figure 2 shows the intensity size distributions obtained for the 2 rice dwarf virus samples, RDV+ and RDV-.
|
The intensity distribution of RDV+ shows a single narrow peak near 75nm diameter. This is relatively close to the size expected from transmission electron microscopy (TEM) data which suggests a diameter of about 70nm. The possible deviation could be due to the fact that in light scattering, the virus is detected in its native environment, and not after being subjected to extensive cooling or other disruptive TEM preparation techniques.
RDV- contains much larger components and suggests that the virus is present as a mix of fragments and oligomers.
The z-average size and polydispersity index (PDI) measured by DLS provide a good indication of the state of the sample. A low polydispersity of PDI<0.1 usually indicates a highly monodisperse sample with only one species present in solution. Higher PDI values are evidence of a broad distribution of different species in the sample. RDV+ showed low polydispersity values throughout the concentration range. The results from DLS are summarized in Table 1.
| RDV+ conc. | z-average | PDI |
|---|---|---|
| 0.031 g/L | 75.2 nm | 0.004 |
| 0.063 g/L | 76.3 nm | 0.037 |
| 0.125 g/L | 76.9 nm | 0.053 |
| 0.250 g/L | 75.9 nm | 0.011 |
| 0.5 g/L | 77.7 nm | 0.050 |
All measurements show homogenous distributions, with an overall z-average of 76.8 ± 1.4nm and a polydispersity index of 0.03 ± 0.02. Sample RDV+ is clearly very monodisperse. The second strain RDV- shows more variation and less homogeneity, as shown in table 2.
| RDV- conc. | z-average | PDI |
|---|---|---|
| 0.031 g/L | 119.2 nm | 0.25 |
| 0.063 g/L | 122.4 nm | 0.24 |
| 0.125 g/L | 117.0 nm | 0.19 |
| 0.250 g/L | 120.8 nm | 0.29 |
| 0.5 g/L | 121.5 nm | 0.28 |
The data indicate very broad size distributions, with an overall z-average of 120 ± 1.9nm and PDI of 0.25 ± 0.04. The observed behavior correlates well with prior studies showing a link between low polydispersity and higher crystallization success rates [2]. DLS may be used to measure dilute samples without knowing the exact concentration.
In static light scattering (SLS), the absolute MW is determined by detecting the scattering signal from known concentrations of the sample. This measurement provides the average molecular weight of all species in solution.
The refractive index increment dn/dc of many proteinsi is approximately 0.175 mL/g [3]. We use this estimate for the virus samples.
The molecular weight obtained was near 80MDa; the second Virial coefficients are 0.00009 and 0.000023 mL mol / g2, for RDV+ and RDV-, respectively. The known MW of the virus is 75MDa. RDV+ behaves as expected, however RDV- has a higher molecular weight because of its higher average size.
| Strain | MW (kDa) | 2nd Virial |
|---|---|---|
| RDV+ | 74400 | 0.000090 |
| RDV- | 86100 | 0.000023 |
Rice dwarf virus (RDV) was characterized by static and dynamic light scattering. The size of the virus is slightly larger than the expected 70nm from TEM, the molecular weight is lower than expected. The sample with lower polydispersity proved to be more suitable for crystallization, the highly polydisperse strain did not form crystals.
[1] A. Nakagawa, N. Miyazaki, J. Taka, H. Naitow, A. Ogawa, Z. Fujimoto, H. Mizuno, T. Higashi, Y. Watanabe and T. Omura et al., "The atomic structure of rice dwarf virus reveals the self-assembly mechanism of component proteins", Structure 11 (2003), pp. 1227-1238.
[2] D'Arcy, A. "Crystallizing proteins - a rational approach", Acta Cryst. D50 (1994), pp. 467-471.
[3] Refractive Increment Data-book: For Polymer and Biomolecular Scientists A. Theisen, C. Johann, M.P. Deacon, S.E. Harding Paperback 64 pages (October 1, 1999) Publisher: Nottingham University Press ISBN: 1897676298
iThe refractive index increment varies between 0.17 and 0.19 mL mol / g2, where large proteins tend to show smaller dn/dc values than small proteins.