The main goal of most Gel Permeation/Size Exclusion Chromatography (GPC/SEC) experiments is to determine the molecular weight distribution (MWD) of the sample or compare molecular weights (MW) of several samples. However, in many cases the MW alone will not explain the differences in sample performance. In the particular case of biologically important molecules such as proteins or polysaccharides, the molecular size may be of equal or even greater importance than the MW to the researcher. Indeed, in the case of proteins, the MW is often known accurately before the experiment and the information required is purely the size of the protein and the proportion and sizes of any associated aggregates. There are two measurements of molecular size in common usage and these are the radius of gyration (Rg) and the hydrodynamic radius (RH). In simple terms, Rg is a mathematically defined dimension describing the distribution of mass centers in the molecule, whereas RH is a phenomenological property of the molecule. This means that for practical purposes, RH is a much more useful measurement, particularly for biologically important molecules.
Direct determination of Rg can only be achieved by measuring the change in scattered light intensity with observation angle. The lower size radius for a multi-angle experiment, even under favorable conditions, is 12-15 nm, as below this size there is no angular dependent information. This means that Rg for almost all proteins and many condensation polymers cannot be measured using the multi-angle technique. At the upper end of the size scale, data can be non-linear, meaning that for many large molecules, such as polysaccharides further information is required to ensure a good fit and extrapolation in order to get an accurate result. A good estimate of Rg can be made from viscometry data without these upper and lower limits by utilizing the Flory-Fox equation linking MW and Intrinsic Viscosity (IV) to Rg.
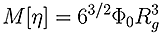
|
The RH can be determined in two ways. The first method is by Dynamic Light Scattering (DLS), which is generally used as a batch technique to measure the average size in the whole sample.
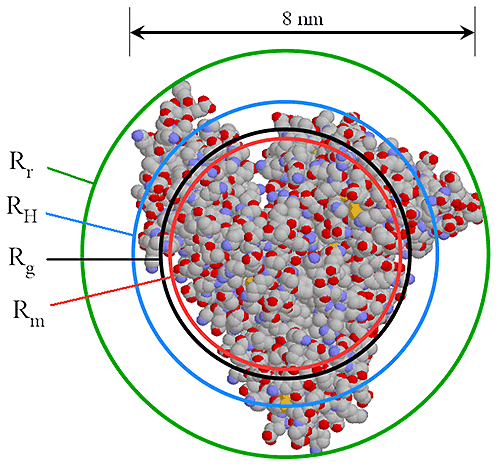
A very accurate, precise and, most importantly, practical method of obtaining RH of all components in a sample after separation is by triple detection SEC. By the use of online light scattering and viscometer detectors, it is straightforward to determine the MW and intrinsic viscosity (IV or [η]) of the polymer or protein at any point on the chromatogram with a high degree of accuracy and precision. This then allows the simple determination of RH at any point. Equation 2 shows the relationship between RH, MW and IV. The figure 1 also shows the relationship between Rh and Rg for BSA.
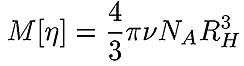
|
|
To demonstrate the use of triple detection SEC in determining accurate molecular sizes, the following instruments and conditions were used. A Viscotek Model 302 triple detector array (TDA) equipped with light scattering, differential viscometer, and refractive index detectors. Two 30 cm mixed bed columns in series. The eluent was 0.5 M LiNO3 flowing at 0.6 mL/min. The samples used were dextran T70 (Pharmacia) at a concentration of ~3 mg/mL with a100 µL injection and bovine serum albumin (Sigma) at a concentration of ~3 mg/mL with a100 µL injection.
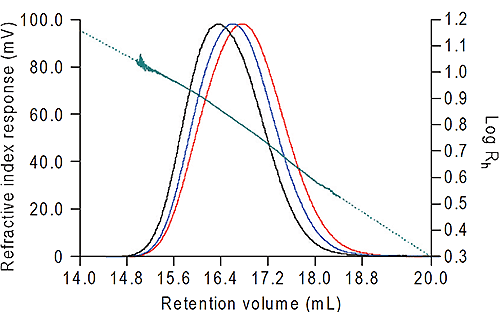
The data were all calculated using Viscotek OmniSEC software. The results from the dextran are shown in figure 2 and from the bovine albumin in Table 1. For the dextran, the Rh was measured across the whole of the peak, ranging from 3.38 nm to 11.03 nm. Note the excellent signal-to-noise on all three detectors which ensures the quality of the size data calculated. At the high end of the distribution the slight curvature in the Rh plot is due to branching in the dextran.
|
The viscometer data can be further utilized to quantify this branching. When the triple detection technique is applied to proteins it can differentiate clearly between the protein and its oligomeric states as shown in Table 1.
| Monomer | Dimer | Trimer | |
|---|---|---|---|
| MW (Da) | 66,400 | 133,000 | 201,000 |
| IV (dL/g) | 0.056 | 0.071 | 0.095 |
| Rh (nm) | 3.88 | 5.32 | 6.69 |
Here the bulk of each peak has been taken to give an average for the monomer, dimer and trimer. The software can give a continuous measurement of size versus molecular weight or retention volume, as in the dextran example but since proteins have discrete molecular weight values for each oligomeric state, there is no continuous distribution. It is clear that triple detection technique provides the most convenient and reliable way to get accurate size values from SEC. It is the only technique which has no upper or lower size limit and is applicable for all molecules that can be separated by SEC.