The work presented here illustrates how Differential Scanning Calorimetry (DSC) is used in the early stages of protein characterization to rapidly provide critical data about protein stability that can be used as a guide to support and accelerate liquid formulation.
Monoclonal antibody development projects exist for a wide variety clinical indications (1,2). Recombinant proteins of commercial interest, including monoclonal antibodies, need additional properties beyond their biological activity to enable their development into successful biopharmaceuticals. Particularly, they need to be resistant to chemical degradation and be physically stable in a given environment, without any tendency to aggregate (3,4). Furthermore, they need a favorable serum half-life and should exhibit no, or very little, immunogenic potential (5).
Apart from being more expensive to produce than liquid formulations, a lyophilized drug product (DP) must be reconstituted by the physician, taking up to 10 to 20 minutes before parenteral administration to the patient. The main driving force pushing the biopharmaceutical industry to focus resources on developing liquid formulations instead of conventional lyophilized DPs is simplified administration. However, there are some technical issues to be overcome with regard to liquid formulations. The main challenge is to keep the protein biopharmaceutical stable in a liquid formulation by maximizing the physical stability and minimizing chemical degradation. It is especially challenging to develop biopharmaceuticals for subcutaneous administration because here the liquid formulation should have as high concentration of protein as possible to compensate for the limitations imposed by possible injection volume, 1.0 to 1.5 ml (6).
|
This places huge pressures on the formulation development teams to deliver finely-tuned liquid formulations that optimally suit each biopharmaceutical. Structure and stability analysis of recombinant proteins, in particular monoclonal antibodies, is of considerable importance and is an indispensable prerequisite for the development of formulations for biopharmaceuticals. A variety of techniques have evolved which can be used to gain structural information and information on protein stability. DSC has become one of the key physicochemical methods to study the stability of protein biopharmaceuticals (7-11). DSC enables the rapid study of protein unfolding without labeling or the use of artificial probes, The technique determines the heat absorbed by the sample as a protein unfolds, giving a measure of its thermostability and an indication of its long-term stability.
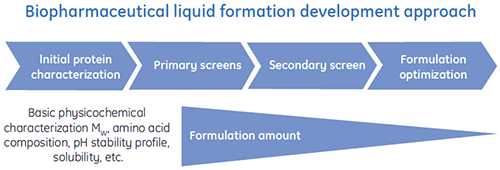
Special liquid formulation development programs have been designed to efficiently deliver optimized liquid formulations for biopharmaceuticals. A general scheme of a typical liquid formulation development process is shown in Figure 1.
DSC has proved to be especially valuable in the early phases of liquid formulation development, a point at which it is preferable to rapidly reduce the number of formulations attempted both to save Drug Substance (DS) and time spent on complex analytics.
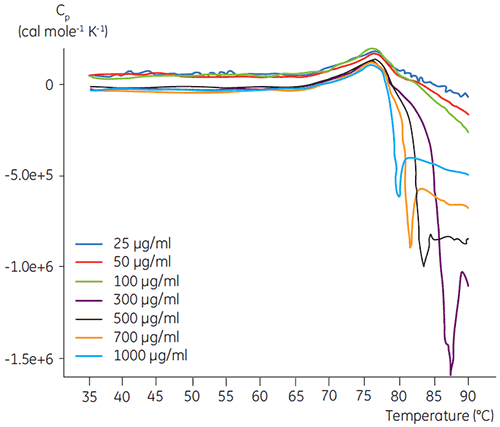
DSC experiments were performed using the Malvern MicroCal VP-DSC (Malvern Instruments) with a scan rate of 1.5 K/min. Samples were degassed for five minutes before analysis. In all experiments, the reference cell of the calorimeter was filled with a buffer corresponding to the sample buffer. Buffer baselines were subtracted from the protein scans and the molar heat capacity was used in the data analysis. The temperature-induced unfolding of all proteins was checked for reversibility by comparing the heating and reheating DSC scans (data not shown). No reversibility was found. The unfolding temperatures were acquired by analyzing the calorimetric profiles according to a two-state transition model. The optimal signal to noise ratio was found when using an antibody concentration of 100 µg/ml (Figure 2). Although higher concentrations did not impact the unfolding temperature, they induced strong exothermic heat due to the aggregation of the protein after unfolding.
|
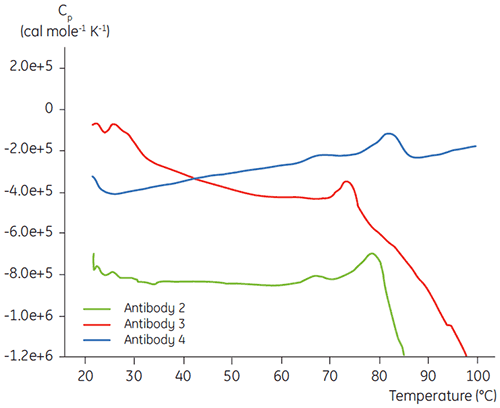
In the initial protein characterization phase of a specific antibody liquid formulation development program, DSC is used for comparing the overall stability of the antibody to other antibodies in development. As can be seen in Figure 3, large differences can be found in unfolding temperatures between different antibodies.
|
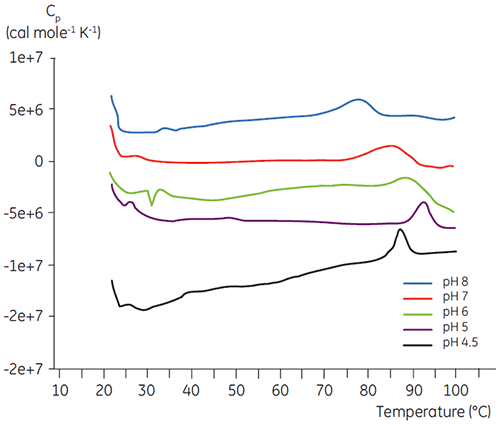
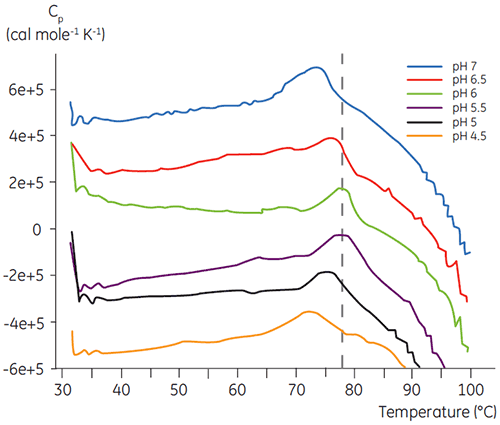
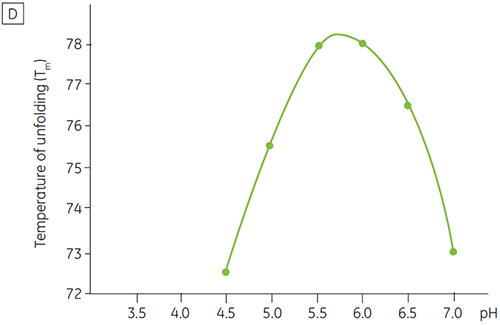
Like most proteins, biopharmaceuticals are very sensitive to pH and it is essential to find the optimal pH to maximize stability. DSC has proved to be an efficient tool for optimizing the pH since information can be gained without performing two and four weeks accelerated stability studies. Although most antibodies typically show the highest stability between pH 5.5 to 6 it is still important to characterize the optimal pH because deviations have been found. Figures 4 and 5 presents the pH profiles of a fusion protein and antibody 2. As can be seen, the optimal pH for the fusion protein (Figure 4) was pH 5 giving a Tm value of 92.8°C. The fusion protein was sequentially destabilized at pH values above pH 5 but was especially sensitive to low pH values. At pH 4, no unfolding could be observed (data not shown). Figure 5 shows a typical pH profile of an antibody. The highest stability was acquired at pH 5.5 and pH 6, giving a Tm value of 78.5°C.
|
|
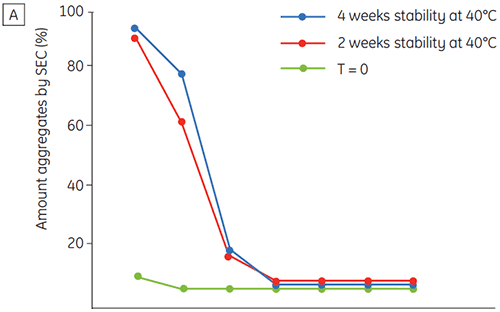
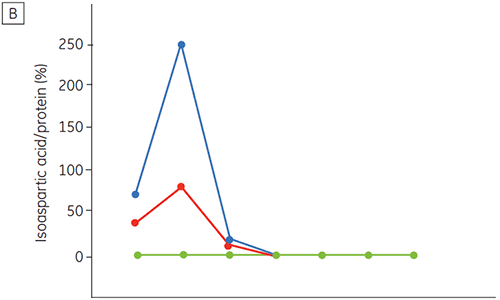
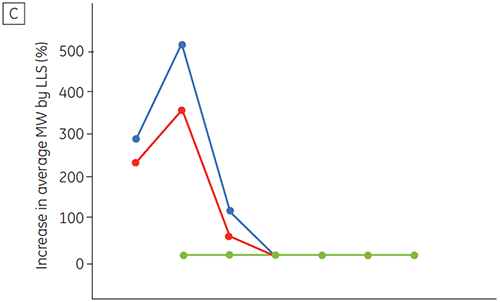
Figure 6 presents data on the chemical and physical stability of antibody 2 as a function of time and pH (initial protein characterization phase). The pH stability profile study shows that after incubating antibody 2 for two and four weeks at ≤pH 5 and at 40°C marked changes in the stability occur. DSC could distinguish the optimal pH condition for this monoclonal antibody by only using the freshly prepared samples (time zero), giving similar results as the other methods (Figure 5). Furthermore, the DSC identified the ≥pH 6.5 values as destabilizing, something that was not obvious in the two and four weeks timeframe but would rather have needed an eight to twelve weeks stability study to be elucidated. In other words the DSC results correspond very nicely with the other methods that were used in this study to optimize the pH of the formulation buffer.
|
|
|
|
The benefit of DSC is that only the time zero samples need to be analyzed to indicate stability. This is because heating the protein is in itself stress inducing. Typically, if the native conformation of a protein is stabilized it can resist thermal stress to a higher degree (unfolds at a higher temperature) than can a protein that is destabilized. Hence, using DSC, it was not necessary to test samples on long term stability at various temperatures simply in order to acquire information about the impact of a specific liquid formulation on protein stability.
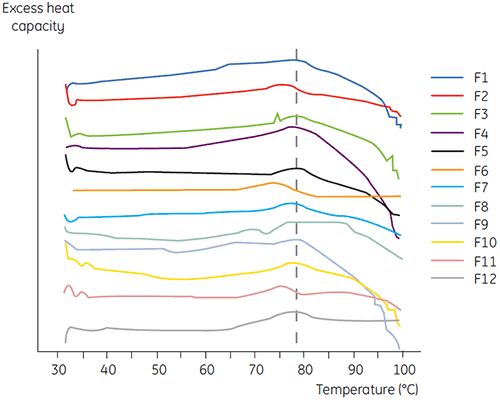
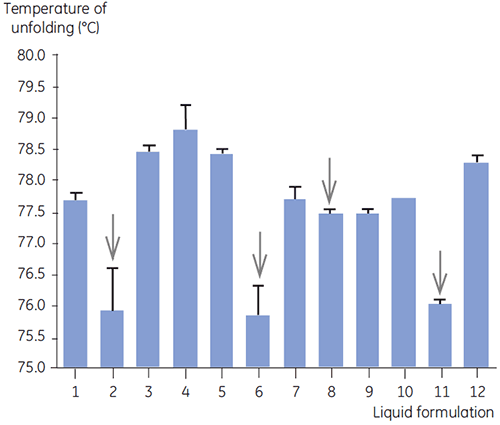
DSC was used in the primary screen of antibody 2 together with other biophysical methods including, SEC, laser light scattering (LLS), capillary electrophoresis (CE), and SDSPAGE, to select the most promising liquid formulations. The samples were put on an accelerated stability study and analysed after four weeks. DSC was found to be especially successful at identifying formulations that had a negative impact on antibody 2, but the technique could also identify good liquid formulations. By analyzing the time zero samples, DSC identified all four liquid formulations (formulation 2, 6, 8 and 11) in the primary screen that were deemed unsatisfactory according to the other analytical techniques. Figure 7 presents thermograms and Figure 8 presents the unfolding temperatures (Tm) of antibody 2 in liquid formulations F1 to F12 from the primary screen. The unfolding of antibody 2 in formulation F9 gave a markedly deviating unfolding pattern. For this reason it was regarded as a critical formulation.
|
|
DSC is a technology that is valuable in the early phases of protein characterization and formulation development. Critical data about protein stability can be rapidly acquired and used as a guide to support and speed up liquid formulation development projects.
This application note was authored by Dr Fredrik Ollila, Novartis Pharma AG.