DSC data was used to identify the most stabilizing elution conditions for the antibody capture step during process development of a protein and the result in cost saving.
Biopharmaceutical process development can be costly and time-consuming. The ultimate goal is to maximize the yield of purified product by the most cost-effective, reproducible and robust route. Since batch failures can have major economic consequences, a thorough understanding of protein stability throughout the development pathway, from research through clinical development to commercial Manufacturing, is fundamental to biopharmaceutical drug commercialization.
Maintaining stability and preserving the active structure of a protein biopharmaceutical until it is administered is essential. This can be accomplished, in part, through stability studies designed at understanding the physical behavior of biotherapeutics. Proteins typically have inherent stability issues because of their complexity and delicate structure. Stability studies to determine the effect of various environmental factors, such as pH, temperature, and ionic strength, on the conformational integrity of a protein, which directly influences biological activity, are indispensable during process development. These stress studies help identify features that are critical by revealing the weak points in the molecule and they provide a rational approach in identifying denaturation mechanisms and in developing effective counter measures, through the selection of appropriate buffer, pH, ionic strength and excipients.
|
Conducting stability studies in parallel with purification method development allows scientists to design more effective methods. During development of purification processes, stability studies can:
Stability studies also aid in the development of robust and stability-indicating assays. These data enable accelerated development of formulations for drug product, by minimizing the range of conditions that need to be explored during typical formulation development studies. This, in turn, translates into resources, cost and time savings.
This application note focuses on the utility of Differential Scanning Calorimetry (DSC) to guide the development of antibody purification processes. In particular, this note describes how DSC data were used to identify optimal elution conditions during protein A capture step development, which resulted in cost savings at the early clinical manufacturing scale.
DSC provides a way to monitor protein stability throughout processing and handling. This can lead to improved conditions that can protect the molecule when atypical conditions or protein behavior occur during processing or assay development.
All proteins were prepared in the indicated buffer by dialysis or buffer exchange with a PD-10 desalting column (GE Healthcare). Final protein concentrations were approximately 1 mg/ml. Proteins were analyzed with the Malvern MicroCal VP-DSC system using a temperature range of 5°C to 90°C, at a scan rate of 1°C per minute. Analyses of thermograms were performed with Origin™ software using either a two-state or a non-two-state model.
Protein A affinity chromatography is commonly used as the initial capture step for monoclonal antibody purification. Protein A binds to the Fc region of many immunoglobulin (Ig) antibody molecules. This binding specificity and selectivity can result in nearly pure product in a single step. Antibody molecules are produced by cell culture and need to be purified from host cell proteins, nucleic acids and cell culture components. Cell-free conditioned cell culture media is applied to the protein A affinity chromatography medium. The antibodies bind to the immobilized protein A at a neutral pH (around pH 7) and eluted with a low pH buffer (for example, citrate buffer at pH 3.5). The eluted antibodies are neutralized with a solution having a high buffering capacity
such as 1 M Tris pH 9, or via desalting.
One issue with the use of protein A affinity chromatography is that antibodies, as well as other proteins, may be unstable at the low pH required for elution. If the protein is unstable at low pH, it can precipitate during or after elution.
Precipitation is usually dependent on protein concentration: high concentration favors aggregation and precipitation. Thus, although most chromatography media can bind at least 20 g of antibody per liter of medium, the stability of the protein at low pH, post elution, becomes the limiting factor for the loading of the medium. Although many antibodies are pH-sensitive, the mechanism of denaturation
and aggregation varies with structure and will require different buffers for optimal stability.
To improve the loading capacity and economics of the protein A chromatography process, the protein needs to be stabilized in the elution buffer. DSC can be used to characterize the stability of the antibody as a function of pH, and determine which additives can improve the protein stability at low pH. In DSC, an increase in the transition temperature (Tm) suggests an increase in protein stability. The protein A chromatography step can be made more economically viable by stabilizing the antibody in the low pH elution buffer, thus increasing the loading capacity.
In a case study, a client wanted to improve the purification process of their antibody (antibody X). Process information indicated that the initial binding capacity was limited to 2 g of antibody X per liter of protein A medium, due to precipitation of the antibody during elution at higher loading. To understand the effect of pH on the stability of antibody X, the protein was prepared in four different buffers: citrate buffer adjusted with Tris to pH 7.0; phosphate buffer pH 7.3; citrate buffer pH 3.5; and citrate buffer pH 5.0. From DSC data (Table 1) antibody X at pH 7.0 and 7.3 showed a higher Tm than antibody at pH 3.5, indicating that the protein was more stable at higher pH. The unfolding onset temperatures were also higher for pH 7.0 and 7.3 compared to pH 3.5. Antibody X at pH 5.0 had a Tm comparable to pH 7.0, and the unfolding onset temperature was lower, suggesting that pH plays a major role in determining unfolding of this antibody.
| Tm of major transition | Onset of unfolding (°C) | |
|---|---|---|
| Citrate-Tris buffer, pH 7.0 | 68.7 | 60.1 |
| PBS, pH 7.3 | 69.5 | 58.5 |
| Citrate, pH 5.0 | 71.5 | 48.2 |
| Citrate, pH 3.5 | 59.3 | 34.1 |
| Citrate + mannitol, pH 3.5 | 64.7 | 41.0 |
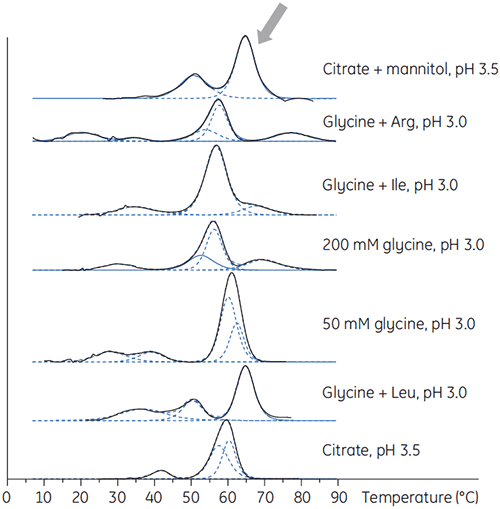
Antibody X was also prepared in low pH buffers with different additives to determine if any of these additives could stabilize the antibody at low pH. DSC experiments were performed to see which conditions would increase Tm of the antibody (Figure 2).
|
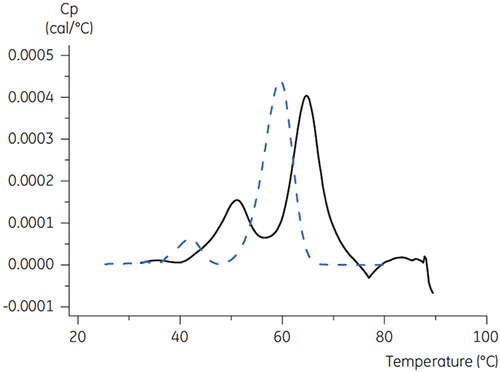
For antibody X, citrate plus mannitol at pH 3.5 resulted in the largest Tm shift, indicating the most favourable stability. The addition of mannitol increased both the Tm and the unfolding onset temperature relative to citrate at pH 3.5. (Figure 3).
|
The DSC results suggested that the addition of mannitol to the elution buffer for the protein A affinity medium would improve the stability of antibody X. Improved antibody stability during elution can increase the loading of the antibody onto protein A chromatography medium.
Process development efforts using the protein stability information determined from DSC did result in an increase antibody X loading during the protein A capture step. Use of citrate plus mannitol as the protein A elution buffer resulted in at least a 7.5-fold increase in capacity to ≥15 g of antibody X per liter of protein A medium, as compared to 2 g per liter with citrate buffer only.
Antibody X after the protein A capture step was more concentrated than it was in the earlier process, eliminating an ultrafiltration/diafiltration step, resulting in savings in materials and processing time.
In this application note, DSC data were used to identify the most stabilizing elution conditions for the antibody capture step during process development of a protein. DSC provides valuable information for choosing protein stabilizing buffering conditions before chromatographic development is undertaken. The ability to improve performance in this critical step in the development of purification processes may result in substantial financial savings downstream..
This application note was authored by Prathima Acharya, formerly with Diosynth Biotechnology. The contribution of Rochelle Bazemore, Sorina Morar, Sue Cook and Jessica Weaver from the Purification Development Group to this work is acknowledged.