In this application note, ITC and DSC enabled characterization of protein-excipient binding as one of the many interactions possible in a complex protein solution formulation.
Biopharmaceutical Technologies, GlaxoSmithKline R&D, 709 Swedeland Rd,
King of Prussia, PA 19406 (USA)
Development of protein-based therapeutics is a challenging task due to inherent protein instabilities, which can manifest as physical instability (unfolding, aggregation, adsorption), and chemical degradation (oxidation, deamidation, cleavage). Such instabilities can lead to reduced activity of the protein or even generation of potentially immunogenic species. One approach to protein stabilization is changing the properties of the solvent in contact with it, which can be accomplished by careful selection of the buffer system, adjustment of pH, and addition of excipients/additives (i.e., creating an optimal formulation).
One important factor in solvent-based stabilization of protein drugs is the choice of appropriate excipients; equally important is the optimization of excipient concentrations that provide extended shelf life while also ensuring highest safety to patient. Thus, a critical part of the protein drug formulation development is the selection of excipients that are suitably soluble and nontoxic, maintain the structural integrity of the protein, enable an acceptable product shelf-life, and preserve the biological activity of the product. These excipients can include amino acids, salts, metals, surfactants, sugars and polyols, and polymers. They can serve as stabilizers, surface-active agents, antimicrobial agents, or antioxidants. Their stabilizing effects are usually protein-specific and concentration-dependent.
Together with the excipient selection, information needs to be gathered not only on optimum excipient concentration, but also on the interactions between various formulation components. Optimizing the selection and concentration of each excipient can be a labor-intensive task that involves extensive formulation screening and stability studies. While general principles of stabilization have emerged from the literature over the past decade (1), the mechanisms by which excipients can improve the stability of a protein during storage are still incompletely understood. Knowledge of the mechanisms of excipient interactions with proteins would eliminate a purely empirical screening approach and allow a rational design and optimization of protein formulation, thus decreasing the time and material requirements for protein product development. Moreover, information on the strength and type of protein-excipient interactions would help predict the protein drug behavior in vivo.
Biophysical analysis techniques have proven to be extremely useful in excipient screening for formulation development. In particular, calorimetry is one of the most efficient methods for assessing protein stability and interactions, as it allows a complete thermodynamic characterization of the system, provided all events are reversible. Calorimetric investigations to explore protein-excipient interactions have been increasingly applied toward the design and optimization of biopharmaceutical formulations. Calorimetry is based on the principle of determining the energetics and stoichiometry of macromolecular interactions by measuring the heat changes resulting from association, dissociation, and/or unfolding processes. ITC is primarily used to determine thermodynamic binding parameters such as binding affinity and dissociation constants, as well as stoichiometry, enthalpy, entropy, and Gibbs free energy of binding under isothermal conditions. It can also be used to determine heat capacity changes upon binding by performing experiments over a range of temperatures. DSC data supplies the thermodynamic parameters of protein unfolding, including the midpoint unfolding temperature, enthalpy, entropy, Gibbs free energy, and heat capacity changes upon unfolding.
These thermodynamic parameters are commonly used to compare protein stability in different formulations, or to determine the relative stabilizing or destabilizing effects of certain excipients.
This application note presents an example of how calorimetry aided the formulation development for ProX by providing insight into excipient-protein interactions. Polysorbate-80 and phenol are examined here as potential additives to the formulation buffer for ProX. Polysorbate-80 is a commonly used surfactant to prevent non-specific adsorption and aggregation of proteins and has been shown previously to interact with proteins (2, 3). It protects the protein against surface-induced aggregation by binding to exposed hydrophobic regions on the protein molecule surface (3). The most commonly used levels of polysorbate-80 are 0.002 to 0.1% (w/v). Phenol is used as an antimicrobial agent for formulations intended to be dosed more than once from the same container. Phenol is toxic and therefore the concentration used in the formulation buffer must be minimized. The most commonly used levels are 0.3 to 0.5% (v/v).
Experiments were performed with a MicroCal™ VP-Capillary DSC. Four hundred (400) μL of each sample and matching buffer were placed in a 96-well plate.
The samples were scanned from 25° to 100°C at a scan rate of 60°C per hour, with 15 minutes equilibration before each scan. Data were analyzed with Origin version 7 software for DSC following buffer subtraction and concentration correction of the protein scan.
Experiments were performed with an MicroCal iTC200. The sample cell was filled with 250 µL of either 10 or 25 mg/mL ProX, and the titrant syringe was filled with 40 µL of either 60 mM phenol or 50 mM polysorbate-80 in the same buffer as ProX. The reference cell was filled with deionized water. For each experiment, 40 µL of titrant was injected into the sample cell in aliquots of 0.2 to 2 µL at a rate of 0.5 µL per second. The following settings were used: stir speed 1000 rpm, reference power 5 to 8 μCal per second, feedback mode/gain set to high, temperature 25°C, initial delay 60 seconds. Control runs of buffer into buffer, titrant dilution, and ProX dilution were subtracted from each experiment. Data were analyzed with Origin version 7 for ITC. Prior to using the samples for ITC runs, the samples were filtered using 0.2 μm low protein-binding syringe filters (Pall Life Sciences), and each of the protein samples was dialyzed into the same buffer used to prepare the excipient solution to ensure matching buffer in the syringe and cell.
Direct binding of excipients with the active ingredient of a pharmaceutical product may affect the product in several ways. Recent studies have demonstrated that commonly used excipients in pharmaceutical formulations can affect the pharmacokinetics of the active drugs (4, 5). Excipients could also potentially alter the protein drug structure or bioavailability, leading to changes in potency. In addition to a possible change in the pharmacokinetics or potency of the active ingredient, excipient/protein interactions may also affect the shelf life or safety of the drug product. Therefore, the protein interaction with formulation excipients needs to be recognized and understood. If direct interaction (i.e. measurable binding) is detected between the protein and the solution excipients, two important features need to be considered while designing the formulation for a protein drug; first, strength and/or reversibility of excipient binding to the protein, which could hinder or totally mask a critical active site on the protein surface following clinical administration, and second, the concentration of the unbound excipient, which has to be sufficient to achieve desired solution properties. Each of these aspects is exemplified below for ProX.
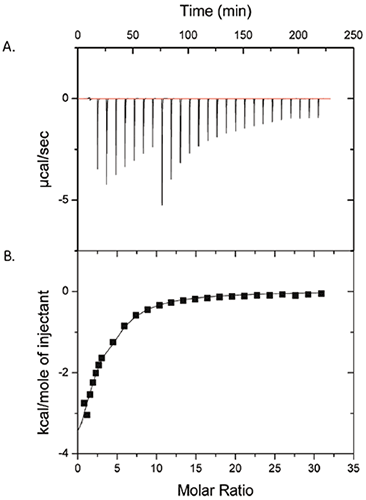
Figure 1 shows the results of an ITC experiment in which 50 mM polysorbate-80 was injected into 25 mg/mL ProX. The raw data are in panel (A), and the integrated heats per injection, after subtraction of the control runs, are in panel (B). The data in panel (B) show that there is indeed a measurable interaction between polysorbate-80 and ProX. The data were fit with a single site model with the binding affinity constant (KA) = 1430 ± 260 M-1, enthalpy of binding (ΔH) = -6.3 ± 1.1 kcal/mol, and number of binding sites (n) per ProX molecule equal to 2.6 ± 0.3. The entropy change of binding (ΔS) was calculated from to be -6.7 ± 3.7 cal/mol-K.
|
Based on the isotherm (Fig 1), it was concluded that the polysorbate-80 binding sites on ProX were saturated at a molar ratio of polysorbate-80 per molecule of ProX of approximately 10. The critical micelle concentration (cmc) of polysorbate-80 is 0.012 mM in pure water and has been reported as high as 0.1 mM in protein solutions (2). The polysorbate-80 concentration in the ITC experiment of Figure 1 spanned 0.1 to 7 mM, above the cmc even after the first injection. This information is important, as any detectable interaction would be attributed to protein binding to the surfactant micelles and not the monomer.
In formulation development, knowledge of the binding parameters is useful for determining the lowest possible concentration of bound excipient needed to 'saturate' the protein and attain the stabilizing effect. Minimizing the concentration of excipient in a formulation reduces costs as well as the level of additives that will be administered to a patient. In this particular case study, a 10-fold molar excess of polysorbate-80 per molecule of protein was sufficient to saturate the protein. The binding parameters may also be used to predict the in vivo behavior of the protein-excipient complex; thus, the weak binding affinity constants measured by ITC suggest that the ProX/polysorbate-80 complex will dissociate due to dilution upon entering the bloodstream with no effect on the biological activity of the protein drug.
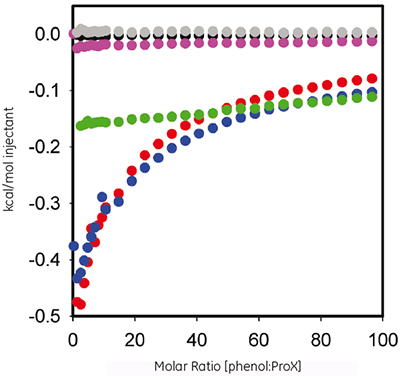
Phenol is used as an antimicrobial agent in ProX formulation. ITC was used here to evaluate the thermodynamics of any measurable binding between the protein and the preservative. Figure 2 plots the heat per injection versus molar ratio for the titration of 60 mM phenol into 10 mgmL ProX at pH 5.7, 4.5, and 3.5, after subtraction of control experiments.
|
The ITC data show that ProX in pH 5.7 and 4.5 interacts with phenol, but not at pH 3.5. The resulting isotherms could not be fitted into any of the pre-defined binding models, indicating that the binding event is complex and involves multiple sites.
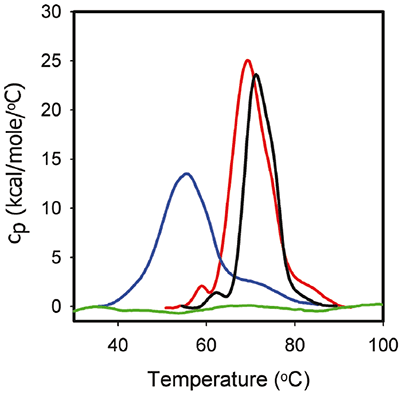
Figure 3 shows the DSC traces for 1 mg/mL ProX in the formulation buffers identical to those used in the ITC experiments. The corresponding DSC data show that, in the absence of phenol at pH 5.7, ProX had at least three unfolding transitions with midpoint unfolding temperatures Tm1, Tm2, Tm3 equal to 59°C, 68°C, and 85°C, respectively, with the middle transition representing the main unfolding event. The total enthalpy of unfolding (ΔHunf) was 270 kcal/mol. At pH 4.5 the first transition was not detectable. The main unfolding event had Tm2 equal to 55°C, and Tm3 decreased to 73°C. The ΔHunf decreased to 216 kcal/ mol, which suggests a decrease in the strength of tertiary interactions. Finally, at pH 3.5, ProX was acid-unfolded as indicated by the absence of any detectable unfolding transitions in the DSC data.
|
Figure 3 also presents the DSC thermogram of ProX unfolding in the presence of phenol at pH 5.7. It is noted that phenol increased the Tm1 by 1°C and Tm2 by 2°C and decreased ΔHunf to 195 kcal/mol. However, the tertiary structure of ProX was the same in the presence and absence of phenol (determined by additional biophysical characterization data such as fluorescence, data not shown). In general, changes in the DSC behavior can be correlated with changes in structure by other methods; however, this is not always the case. The changes in DSC are believed to originate from the disruption of the forces stabilizing the native protein structure (such as van der Waals, hydrophobic and electrostatic interactions, hydrogen bonds, etc.) and the hydration of the exposed residues. Destabilization of the tertiary interactions could decrease the compactness of the protein, which is detected by DSC, without inducing structural changes that are detectable by other analytical methods.
This combination of ITC and DSC data revealed significant information about the phenol-protein interactions. First, phenol binds to folded ProX, but does not bind to unfolded ProX (Fig 2); it is therefore important for tertiary structure to be intact for ProX to have binding sites available for phenol. Second, since it is critical that protein tertiary structure be intact for function, pH 5.7 appears to be the optimum formulation pH. Third, as seen in Figure 2, binding of phenol to ProX reaches full saturation at molar ratios higher than 50. Fourth, the DSC testing (1 mg/mL ProX in pH 5.7 formulation buffer containing 0.005% (v/v) phenol) was performed at a calculated phenol : ProX molar ratio of 38-which was close to, but not at, full saturation. Fifth, although the binding sites were not saturated, this molar ratio of phenol was sufficient to increase the thermal stability of ProX.
Phenol was found to have a decreased antimicrobial activity in the presence of ProX (data not shown). This observation, combined with the knowledge that phenol binds to ProX, indicates that bound phenol has either no, or reduced, antimicrobial activity compared to unbound phenol. With the aid of the binding curve, a concentration of phenol above the saturation-binding point could be selected for the formulation, where the concentration of unbound phenol would allow effective antimicrobial activity. The increased Tm values for ProX in the presence of phenol were not initially predicted; however, it indicates improved thermal stability and suggests better long-term stability of the protein. No direct correlation can be made between phenol-induced stabilization, and the antimicrobial effectiveness of phenol.
Using ITC and DSC, the binding of phenol to folded ProX at pH of 5.7, and of polysorbate-80 to ProX were identified and thermodynamically characterized. The measured binding constant for ProX to polysorbate-80 classifies the interaction as weak; thus, in vivo dissociation of the active protein from the stabilizing excipients in the formulation could be predicted, along with minimal excipient-induced interference with the full biological effects of the protein drug. Based on the saturation curve of ProX-phenol titrations, a concentration of phenol above the saturation point can be identified in order to create a formulation having optimal antimicrobial capability and improved thermal stability. Not only was the hypothesized excipient binding confirmed, but also the suitable concentrations of these two excipients (a 10-fold molar excess of polysorbate-80 per molecule of ProX at concentrations higher than cmc, and a 50-fold molar excess of phenol per molecule of ProX) were identified.
Requirements for protein product formulations include safety and efficacy. Addition of excipients for the purposes of creating safe and efficacious protein therapeutics, as well as, enhancing product stability and shelf life requires the understanding of the interactions between the protein and the excipients at a molecular level. In this application note, ITC and DSC enabled characterization of protein-excipient binding as one of the many interactions possible in a complex protein solution formulation.
Arakawa, T., et al, Factors affecting short-term and long-term stabilities of proteins. Adv. Drug Deliv. Rev. 46, 307-326 (2001).
Chou, D. K., et al, Effects of Tween 20 and Tween 80 on the stability of Albutropin during agitation. J. Pharm. Sci. 94, 1368-1381 (2005).
Bam, N. B., et al, Tween protects recombinant human growth hormone against agitation-induced damage via hydrophobic interactions J. Pharm. Sci. 87, 1554-1559 (1998).
Ren, X., et al, Pharmaceutical excipients inhibit cytochrome P450 activity in cell free systems and after systemic administration. Eur. Jour. Pharm. Biopharm. 70, 279-288 (2008).
Taheri-Kafrani, A., et al, T. Beta-lactoglobulin structure and retinol binding changes in presence of anionic and neutral detergents. J. Agric. Food Chem. 56, 7528-7534 (2008).