The physical properties and behaviour of proteins in solution depend on many factors to do with the purification and formulation of the protein as well as its own innate properties. Size-exclusion chromatography (SEC) is a powerful tool that is commonly used to look at factors such as recovery, molecular weight and aggregation of proteins.
The principle of SEC involves separating a sample as it travels through a porous but inert column matrix. While smaller molecules penetrate the pores more deeply, larger molecules are excluded and thus travel through the column faster. The result is a separation based on hydrodynamic volume but the desire is usually to know the molecular weight. Previously, molecular weight was estimated by comparing the elution time of an unknown protein to that of standard globular proteins of known molecular weight which we term 'conventional calibration'. This was done using a single concentration detector such as ultraviolet (UV). Now, however, the use of a light scattering detector in combination with a concentration detector (UV or refractive index, RI) allows protein molecular weight to be measured independently of retention time. This is a very useful development as many proteins do not have globular structures making their measured molecular weights inaccurate. The addition of further detectors such as a second concentration detector, intrinsic viscosity (IV) and dynamic light scattering (DLS) can greatly increase the amount of information available from a single SEC measurement.
The Malvern SEC-MALS 20 (figure 1) system is a 20 angle light scattering device capable of making measurements of protein molecular weight independent of elution volume. In this application note, a number of proteins are separated using SEC. Their molecular weight is measured by multi angle light scattering (MALS), or by conventional calibration. The differences in results are discussed..
Figure1: The Viscotek SEC-MALS 20 system
|
The SEC-MALS 20 system was connected to a Viscotek TDAmax system using the TDA RI detector to measure concentration. Samples were separated along 2 x Viscotek protein columns using phosphate-buffered saline as the mobile phase. All of the protein samples were dissolved in the mobile phase. The SEC-MALS 20 system was calibrated using bovine serum albumin, a protein with a well characterised molecular weight. Column calibration was performed with a series of globular proteins (Gel Filtration Markers Kit for Protein Molecular weights 29,000-700,000 Da, Sigma-Aldrich).
The detectors and columns were all held at 30°C to ensure a good separation and maximise baseline stability of the detectors.
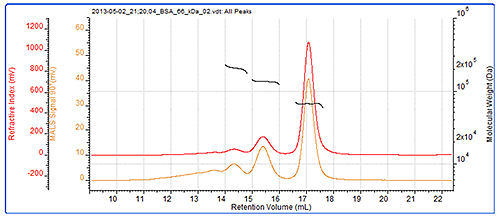
As a system verification, it is always a good idea to check the calibration and the BSA dimer (which is often present in BSA samples) is an excellent verification standard. In this case, the dimer molecular weight has been correctly measured, and even the trimer present in the sample has been measured to a high level of accuracy. The BSA chromatogram can be seen in figure 2 and the results from the BSA measurement can be seen in table 1. Figure 2 also shows the SEC-MALS chromatograms for the BSA. As an isotropic scatterer, the peak sizes and detector responses are the same at each angle. The molecular weights across all of these peaks are clearly seen to be very stable. This is expected with proteins which individually have very tightly controlled molecular weights and are considered to be monodisperse. This and the molecular weight values indicate that these peaks are different oligomers of the BSA.
|
|
| Trimer | Dimer | Monomer | |
|---|---|---|---|
| Peak RV - (ml) | 14.31 | 15.37 | 17.05 |
| Mn - (kDa) | 203.8 | 135.2 | 66.4 |
| Mw - (kDa) | 204.4 | 135.3 | 66.5 |
| Mw/Mn | 1.003 | 1.001 | 1.001 |
| Wt Fr (Peak) | 0.054 | 0.165 | 0.78 |
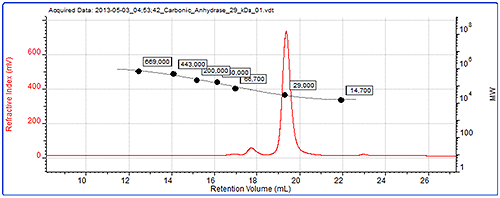
Figure 4 shows the data from the conventional calibration. The chromatogram shows refractive index signal for carbonic anhydrase, one of the standards and the generated calibration curve.
|
The molecular weights of the BSA dimer and trimer were measured by conventional calibration and the results are shown in table 2 and can be compared with the SEC-MALS data in table 1. Since the structure of these oligomers is no longer as globular as the monomer, their molecular weights are not accurately calculated using column calibration. In the absence of light scattering, there is no way to know whether the molecular weight measured using column calibration is accurate. As such, these peaks could mistakenly be identified as trimer and pentamer. However, the light scattering results (table 1) can be used to correctly identify the dimer and trimer through accurate measurement of their molecular weight.
| Trimer | Dimer | |
|---|---|---|
| Peak RV - (ml) | 14.32 | 15.33 |
| Mn - (kDa) | 341.1 | 198.4 |
| Mw - (kDa) | 345.1 | 202.2 |
| Mw/Mn | 1.012 | 1.019 |
| Wt Fr (Peak) | 0.021 | 0.103 |
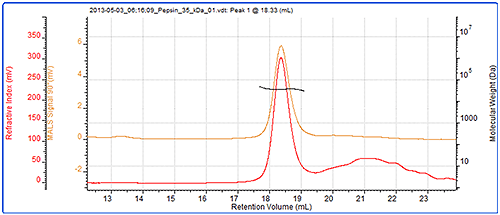
Finally, the molecular weight of another protein, pepsin has been measured by SEC-MALS and column calibration. The chromatogram and light scattering results are shown in figure 5. Table 3 shows a comparison of the measured molecular weights.
|
| MALS | Column calibration | |
|---|---|---|
| Peak RV - (ml) | 18.33 | 18.63 |
| Mn - (kDa) | 34.6 | 38.5 |
| Mw - (kDa) | 34.7 | 39.6 |
| Mw/Mn | 1.004 | 1.028 |
Pepsin's molecular weight has been correctly measured at 35 kDa by SEC-MALS but incorrectly measured at 40 kDa by column calibration. The reason for this is that the structure of pepsin is not globular and is it therefore larger than would be expected for a globular protein. It therefore elutes earlier and records a higher molecular weight by conventional calibration than is correct. On the other hand, the molecular weight from light scattering is independent of retention volume and is therefore measured correctly.
This application note has shown the successful measurement of the molecular weight of a number of proteins using the Malvern SEC-MALS 20 system. The addition of MALS to an SEC system allows the measurement of protein molecular weight independent of a protein's elution volume or structure. Pepsin and BSA oligomers were both well characterised by the SEC-MALS system, as their molecular weights were correctly measured.