Proteins are composed of polypeptide chains, synthesized within the cell from a pool of 20 different amino acid types. In contrast to manmade and random coil biological polymers, the protein's polypeptide chains are folded into unique 3-dimensional structures in the natured state. These structures are stabilized by a combination of electrostatic and hydrophobic interactions, along with the formation of multiple hydrogen bonds between the side chains of the amino acids within the structure.
Proteins were evolved to perform specific functions within biological systems, such as catalysing reactions and transporting small molecules or ions. This functionality necessitates a large degree of flexibility within the structure of the protein. As such, the interactions stabilizing the protein structure are just sufficient to maintain the structure within a narrow range of environmental conditions. If solution conditions are outside of this range, e.g. through a change in the pH or temperature, entropically driven denaturation or unfolding can quickly occur. When denaturation occurs, the small size of the protein is increased to a value consistent with a random coil polymer of the same molecular weight. In the absence of chaotropic (aggregation prohibiting) agents, inter-polymer hydrophobic interactions can quickly lead to non-specific aggregation of the denatured polypeptide chains.
A representation of the denaturation process, along with the subsequent change in size is shown in Figure 1.
|
Dynamic light scattering (DLS) has long been used as a protein characterization tool. In DLS, the Brownian motion of the particles is measured, and the hydrodynamic size is calculated using the Stokes-Einstein equation. The sensitivity of DLS is sufficient to distinguish different oligomeric and quaternary protein states, and is ideally suited for monitoring the stability of the protein structure to denaturing conditions.
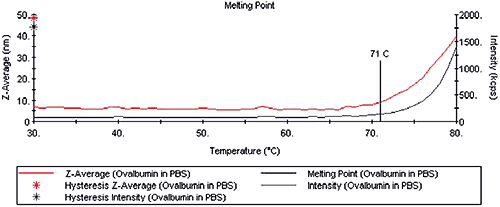
The protein melting point (TM) is defined as the temperature at which the protein denatures. The change in size that accompanies the protein denaturation is easily identified using DLS techniques. Consider Figure 2 for example, which shows the temperature dependent Z-Average diameter and scattering intensity for Ovalbumin in phosphate buffered saline (PBS) measured with a Zetasizer Nano ZS. At temperatures less than 71°C, the size and scattering intensity are constant, suggesting a stable tertiary structure. At 71°C and higher, both the size and scattering intensity increase exponentially with temperature, indicating the presence of denatured aggregates.
|
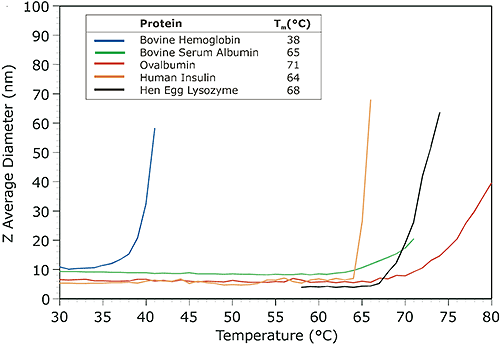
Because of the unique primary sequence of amino acids associated with specific protein types, melting points can differ significantly. Solutions conditions, e.g. pH and salt concentration, and post translational modifications, e.g. glycosylation, can also have a marked influence on the stability of the protein structure and hence the melting temperature. Melting point measurements then are an important step in the total characterization of the target protein, particularly if the protein is destined for integration into a pharmaceutical product or formulation. Figure 3 shows the melting point traces for a series of proteins in PBS. The automated measurements were collected with a Zetasizer Nano ZS, using a 1°C incremental temperature ramp and a 3 minute equilibrium time at each measurement temperature.
|
The Zetasizer Nano ZS from Malvern Instruments is the first commercial instrument to include the hardware and software for combined dynamic, static, and electrophoretic light scattering measurements, giving the researcher a wide range of sample properties, including the size, molecular weight, and zeta potential. The system was specifically designed to meet the low concentration and sample volume requirements typically associated with pharmaceutical and biomolecular applications, along with the high concentration requirements for colloidal applications. Satisfying this unique mix of requirements was accomplished via the integration of a backscatter optical system and the design of a novel cell chamber. As a consequence of these features, the Zetasizer Nano ZS specifications for sample size and concentration exceed those for any other commercially available DLS instrument, with a size range of 0.6nm to 6um, and a concentration range of 0.1mg/mL lysozyme to 40% w/v. As an added bonus, the Zetasizer Nano ZS hardware is self optimizing, and the software includes a unique "one click" measure, analyze, and report feature designed to minimize the new user learning curve.